Why Spatial Transcriptomics?
Spatial information is critical for understanding gene expression patterns within tissue and how individual cells interact with their surrounding environment.
In recent years, single-cell technology has gained favor over traditional bulk RNA approaches. Bulk RNA sequencing masks heterogeneity within a sample, which makes it difficult to characterize complex tissue such as tumors. Single-cell RNA sequencing allows for identification of rare or unique cell populations that are otherwise difficult or impossible to isolate.
However, today’s single-cell technologies require dissociation of tissue and subsequent loss of spatial information. Spatial transcriptomics allows you to acquire gene expression data at single-cell or near-single-cell resolution without losing spatial information.
Spatial transcriptomics methods are now being used to identify biomarkers and elucidate the mechanisms underlying disease. Spatial transcriptomics datasets can be integrated with other single-cell experiments to better characterize complex tissues.
How it works
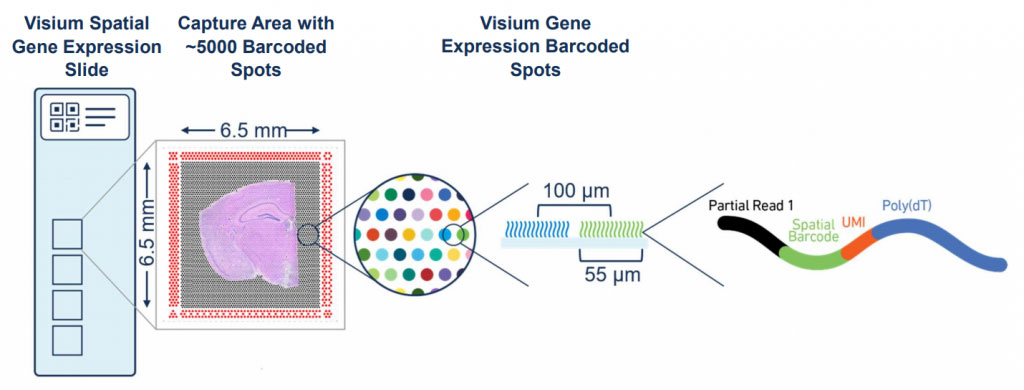
At Three Dimension Genomics, we use Visium, a whole-transcriptome spatial method developed by 10X Genomics. The Visium Spatial Gene Expression slide has 4 captures areas that each span 6.5mm x 6.5 mm and hold ~5,000 barcoded spots. Since an individual spot is 55um, you can expect to capture between 1-10 cells per spot depending on your tissue type. Tissue sections (either fresh-frozen or FFPE) are cut and placed onto the 10X Visium slide, which is then H&E stained (or stained for immunofluorescence) and imaged with a brightfield microscope. The spot’s spatial barcode is retained throughout library preparation and later used to visualize gene expression across the tissue.
Advantages of 10X Visium
10X Visium is compatible with both fresh-frozen and FFPE samples and has been optimized across a variety of human and animal tissues. When profiling fresh-frozen tissues, Visium is suitable for any species with a reference genome.
10X Visium has high resolution compared to other spatial transcriptomics technologies, allowing for identification of 1-10 cells per barcoded spatial area. It also has advantages over in situ hybridization-based approaches (such as single-molecule FISH), which require targeting to a limited number of mRNAs. 10X Visium, however, allows unbiased detection of mRNAs across the whole transcriptome.
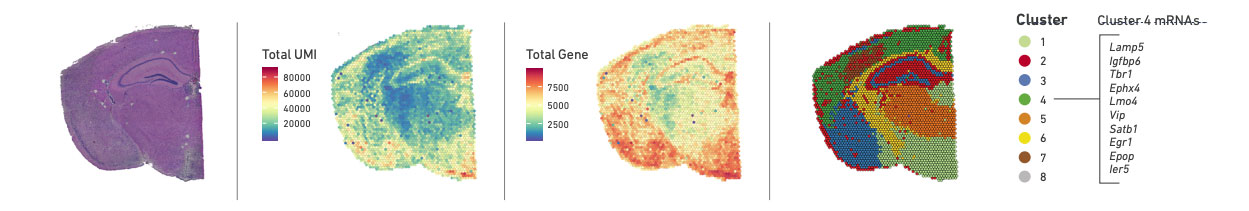
10X Genomics provides an interactive software called Loupe Browser that allows for easy navigation of Visium data. You can visualize the spatial distribution of UMI counts, gene counts, and identified spot clusters, as well as expression of individual genes of interest.
Spatial transcriptomics + single-cell RNA-seq: a powerful combination
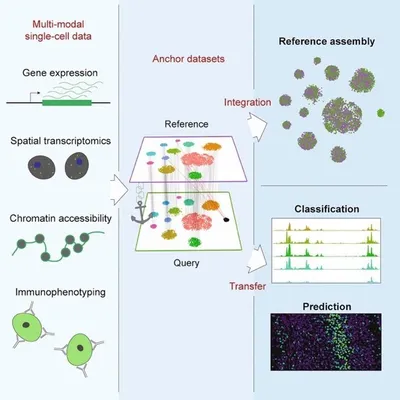

When performing a spatial transcriptomics experiment, we recommend running a single-cell or single-nuclei RNA sequencing study in parallel. This is typically done using serial sections of the same tissue block. This provides several advantages.
Better identification of cell types. In a Visium experiment, each spot may capture up to 20 cells, and the spot RNA may be thought of as a “bulk RNAseq” population representing a small mixture of heterogeneous cells. Deconvoluting the spot transcriptome into separate cell types is typically done using a reference set of publicly available single cell RNAseq data for cell types that are believed to be in the tissue of interest. However, the quality and validity of cell type assignments is greatest when the single cell or single nuclei RNAseq data are obtained from the same Visium tissue sample. For example, Visium + single cell/single nuclei profiling of serial sections can capture rare or novel cell types that may be present in your tissue but missing from reference datasets.
Better detection of genes. Single-cell RNA-seq by definition captures individual cells and at greater sequencing depth per cell than is possible with spatial transcriptomes. Integration of single cell and spatial transcriptomes boosts average read, UMI and gene counts per cell providing more in-depth information from your spatial experiments.